Mike has also created a number of Excel tables which have a lot of configuration changes ect in them. This is a listing of some of the useful files. The Event Logs section on the time series page gives you a more complete listing.
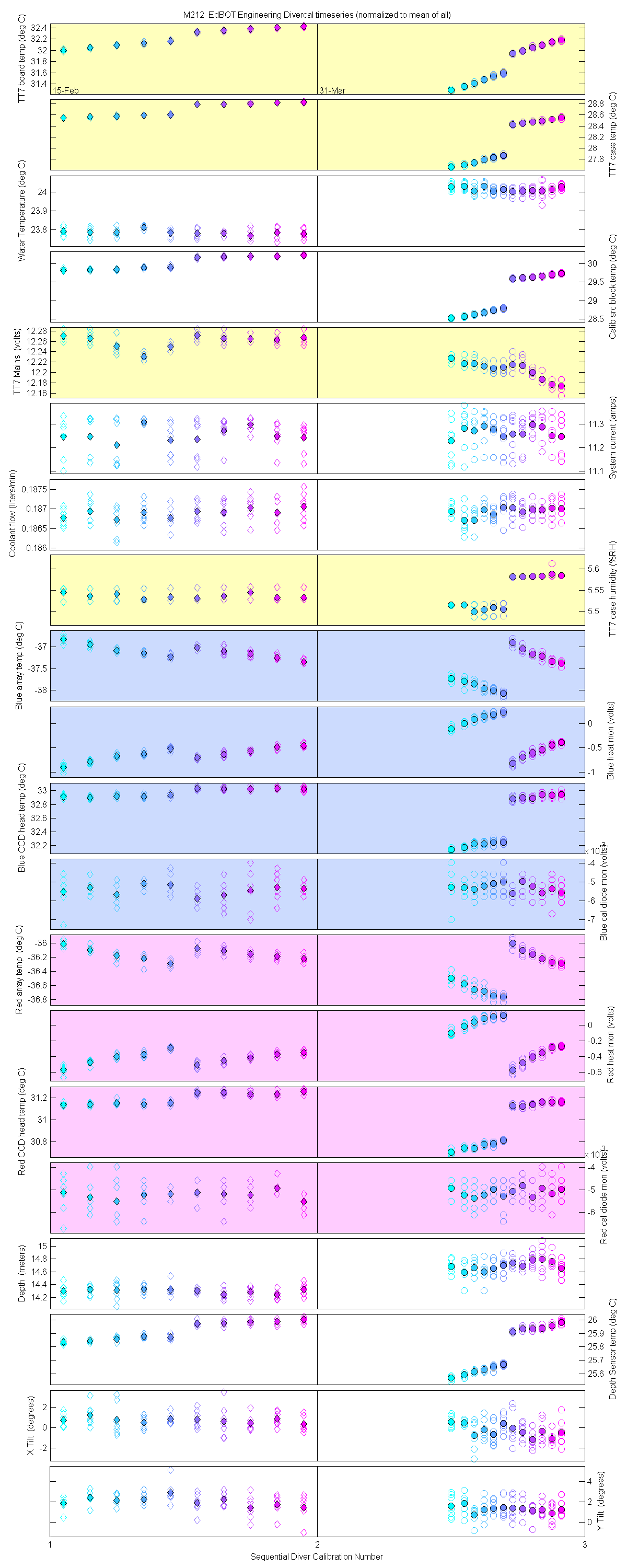
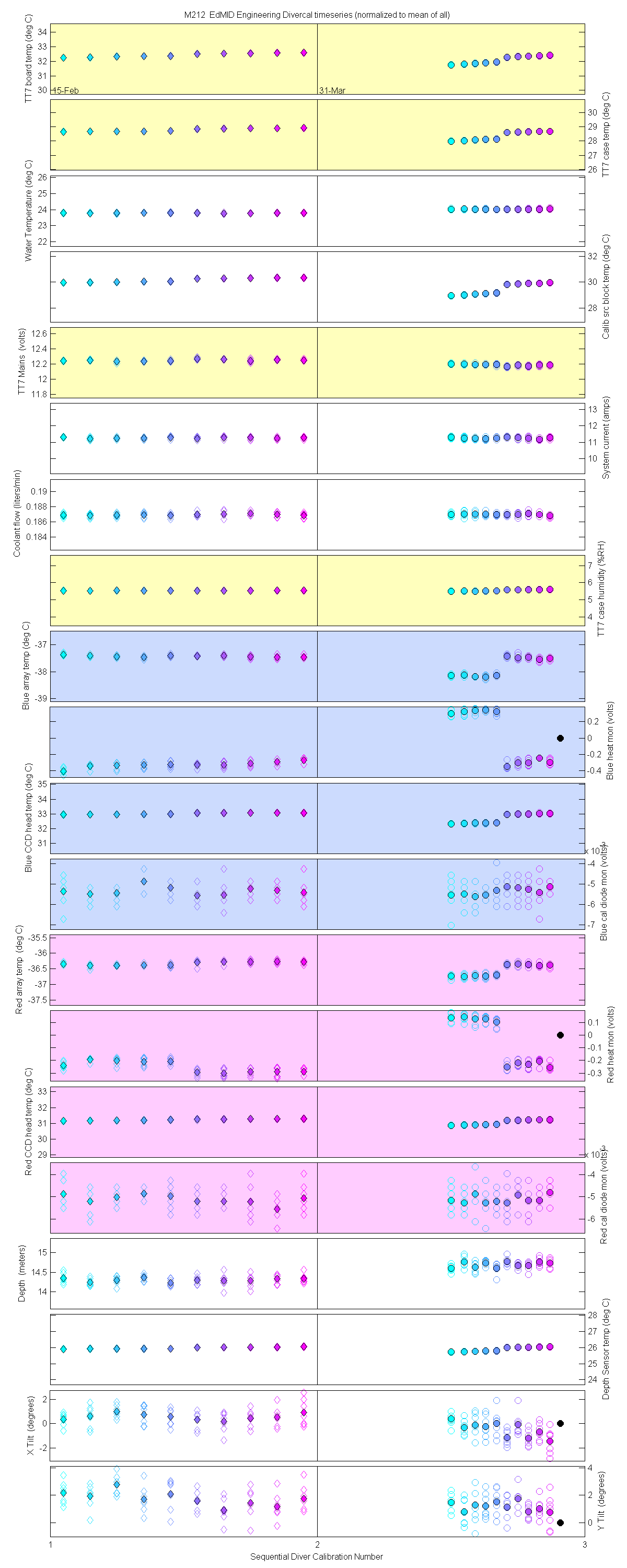
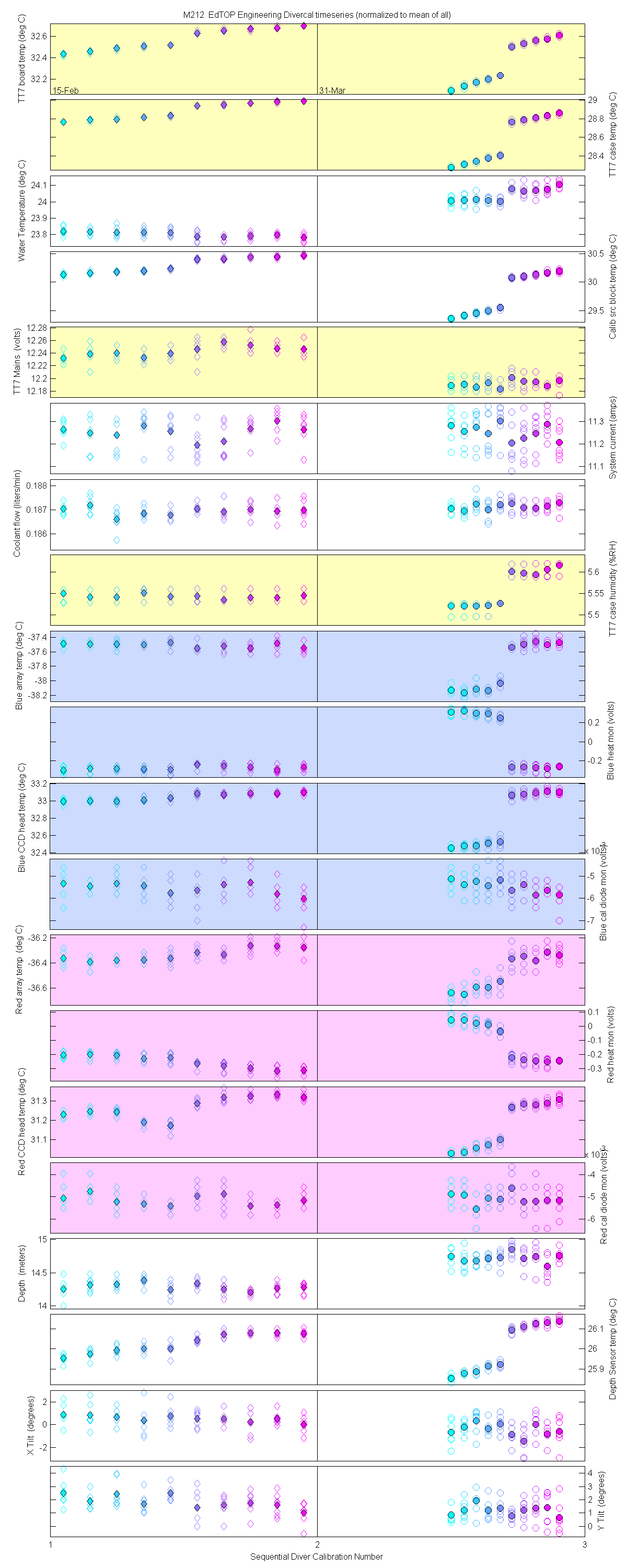
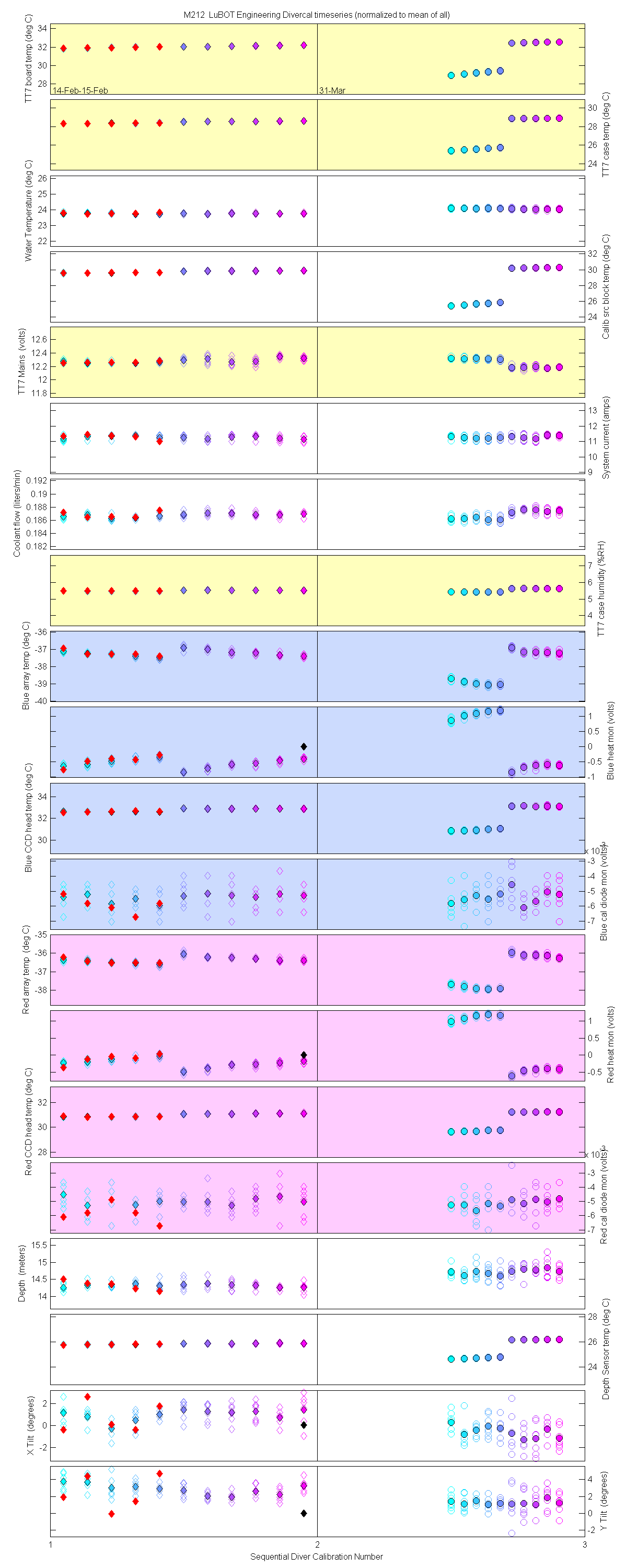
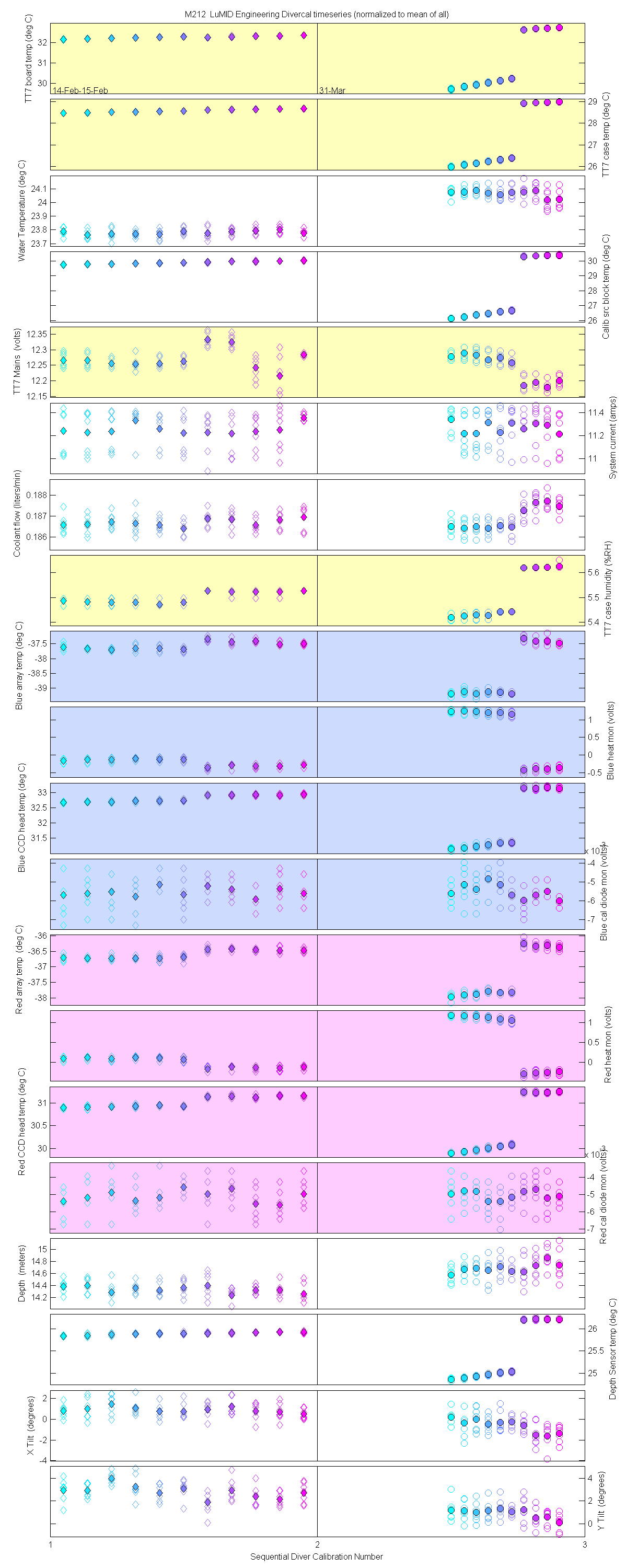
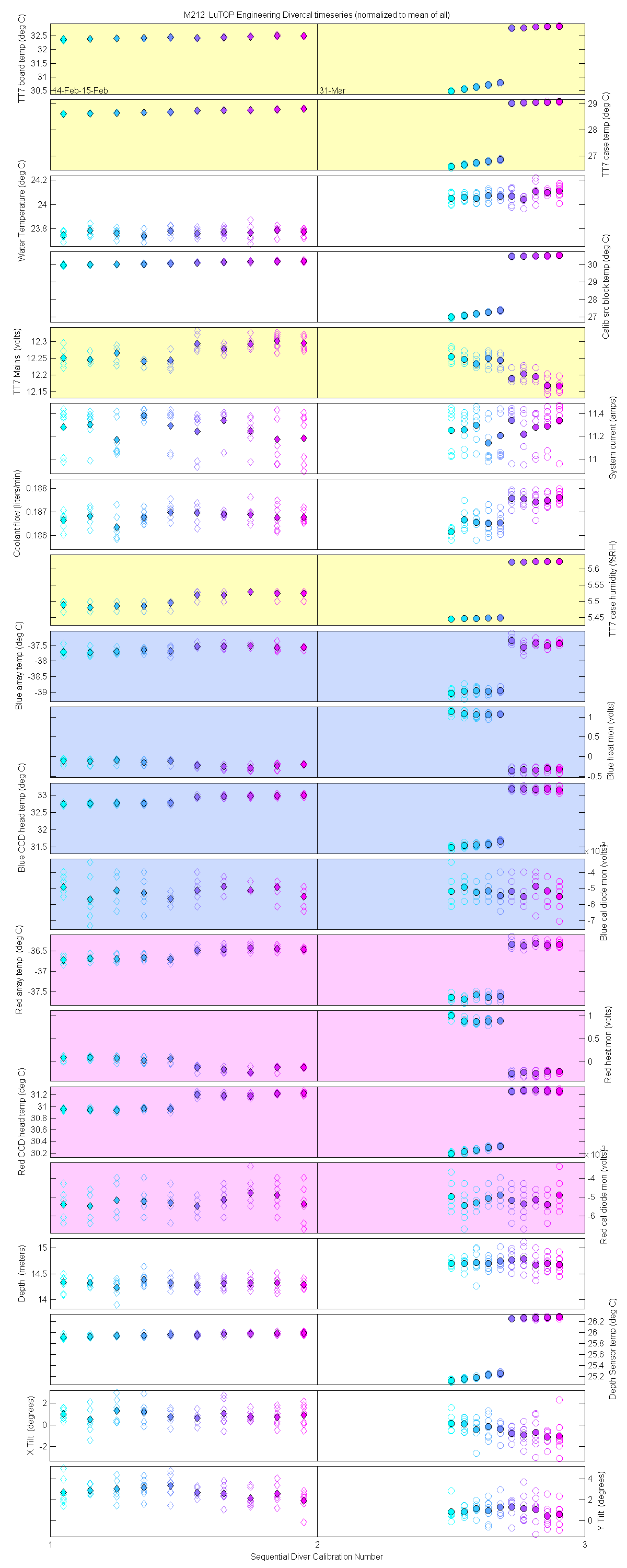
Plots of the auxilliary data for each sensor are at the bottom of this page
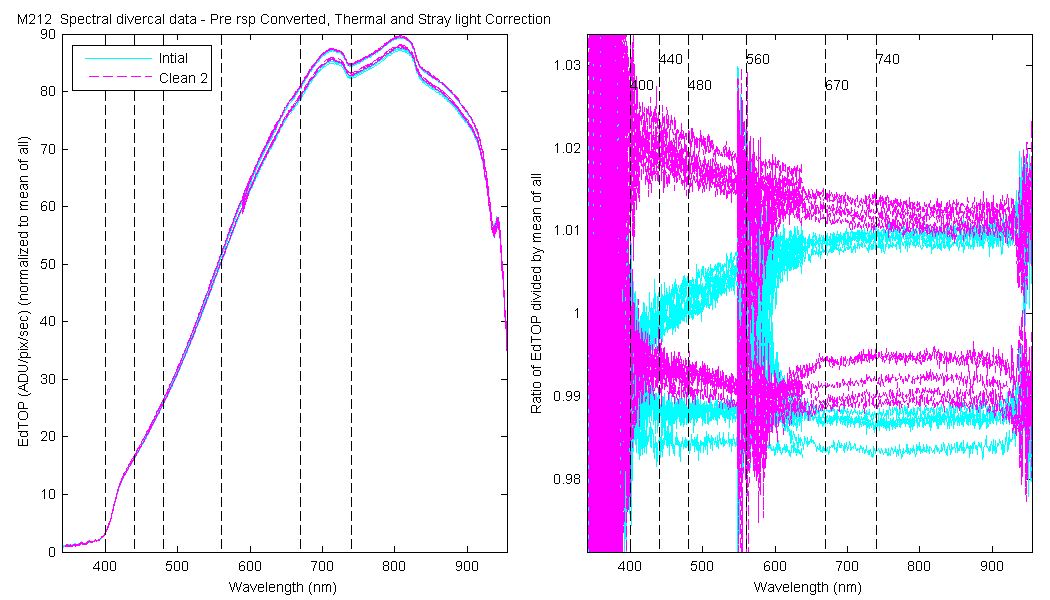
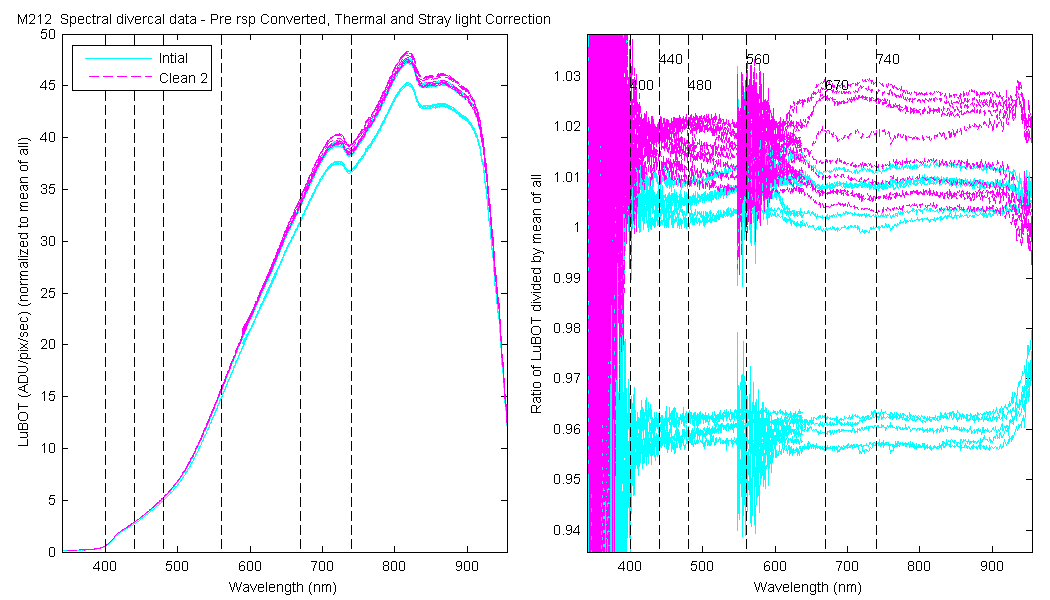
NOTE: All the data startes in units of ADU/pix/sec and is thermally and stray light corrected and the data are converted.
| Cruise Name | Dates | Details | LOG & IMG |
|---|---|---|---|
| MOBY-L54 | 10-Feb-2000-15-Feb-2000 | MOBY M211 and the old mooring buoy were retrieved on 10 Feb (afternoon), and the new mooring buoy was deployed on 11 Feb. MOBY M212 was successfully deployed on 13 Feb. Intercomparison data were also collected on 13 Feb, coincident with the MODIS overpass (20:30 GMT), for Rads-UW, MOS, SPMR, TAPS, and MOBY M212. Initial M212 diver cals were performed by Hawaiian Rafting Adventures personnel on 14 Feb. | IMG |
| MOBY-L55 | 29-Mar-2000-31-Mar-2000 | Maintenance and diver calibrations of MOBY212: On 29 March, Mark Yarbrough, Mike Feinholz, Yong Sung Kim, and HRA personnel replaced the meteorological stations controller box and anemometer. The ARGOS battery was replaced, and the WARS camera was tested on 30 Mar. Diver calibrations were performed on MOBY212 radiance and irradiance sensors on 31 Mar. | IMG |
| MOBY-L56 | 15-May-2000-18-May-2000 | MOBY exchange via R/V Kaimikai-O-Kanaloa: On 15 May MOBY213 was successfully deployed in the morning and tethered to the mooring and MOBY212 was placed under tow behind the ship for the night. The two MOBYs made coincident optical profiles on 16 May before MOBY212 was recovered. Intercomparison data were collected on 17 and 18 May with MOS, SPMR, FOS, WARS, and MOBY213. Mark Yarbrough worked with Hawaiian Rafting Adventures (HRA) on 17 and 18 May to service the CIMEL site and perform diver calibrations on MOBY213. | IMG |
| MOBY-L57 | 19-Jun-2000-21-Jun-2000 | Diver calibration via HRA: CIMEL #93 was removed from Lanai on 19 Jun. Diver cals were successfully performed 20 - 21 Jun. | IMG |
| Sensor | Re-Process Code | 400.0 nm | 440.0 nm | 480.0 nm | 560.0 nm | 670.0 nm | 740.0 nm |
|---|---|---|---|---|---|---|---|
| LuTOP | Mean(Pre & PostCal) | 1.077 | 1.063 | 1.054 | 1.044 | 1.038 | 1.036 |
| LuMID | Mean(Pre & PostCal) | 1.037 | 1.027 | 1.018 | 1.009 | 1.009 | 1.006 |
| LuBOT | Mean(Pre & PostCal) | 0.987 | 1.008 | 1.013 | 1.013 | 1.006 | 1.003 |
| EdTOP | Mean(Pre & PostCal) | 0.975 | 0.979 | 0.982 | 0.983 | 0.993 | 0.994 |
| EdMID | Mean(Pre & PostCal) | 1.128 | 1.115 | 1.106 | 1.092 | 1.079 | 1.074 |
| EdBOT | Mean(Pre & PostCal) | 1.037 | 1.041 | 1.039 | 1.035 | 1.029 | 1.027 |
The above table shows the ratios of the pre/post system response (if a post exists) and the Re-Process Code indicates if the system response is a mean of the pre and post system, or made only from the pre or post calibration.
| Diver Cal # | Type | Lu Scan Sets |
Ed Scan Sets |
Eu Scan Sets |
Filenames |
|---|---|---|---|---|---|
| 1 | Initial | 32 | 30 | 0 | 00021423 00021500 00021501 |
| 2 | Clean | 30 | 32 | 0 | 00033119 00033120 00033121 |
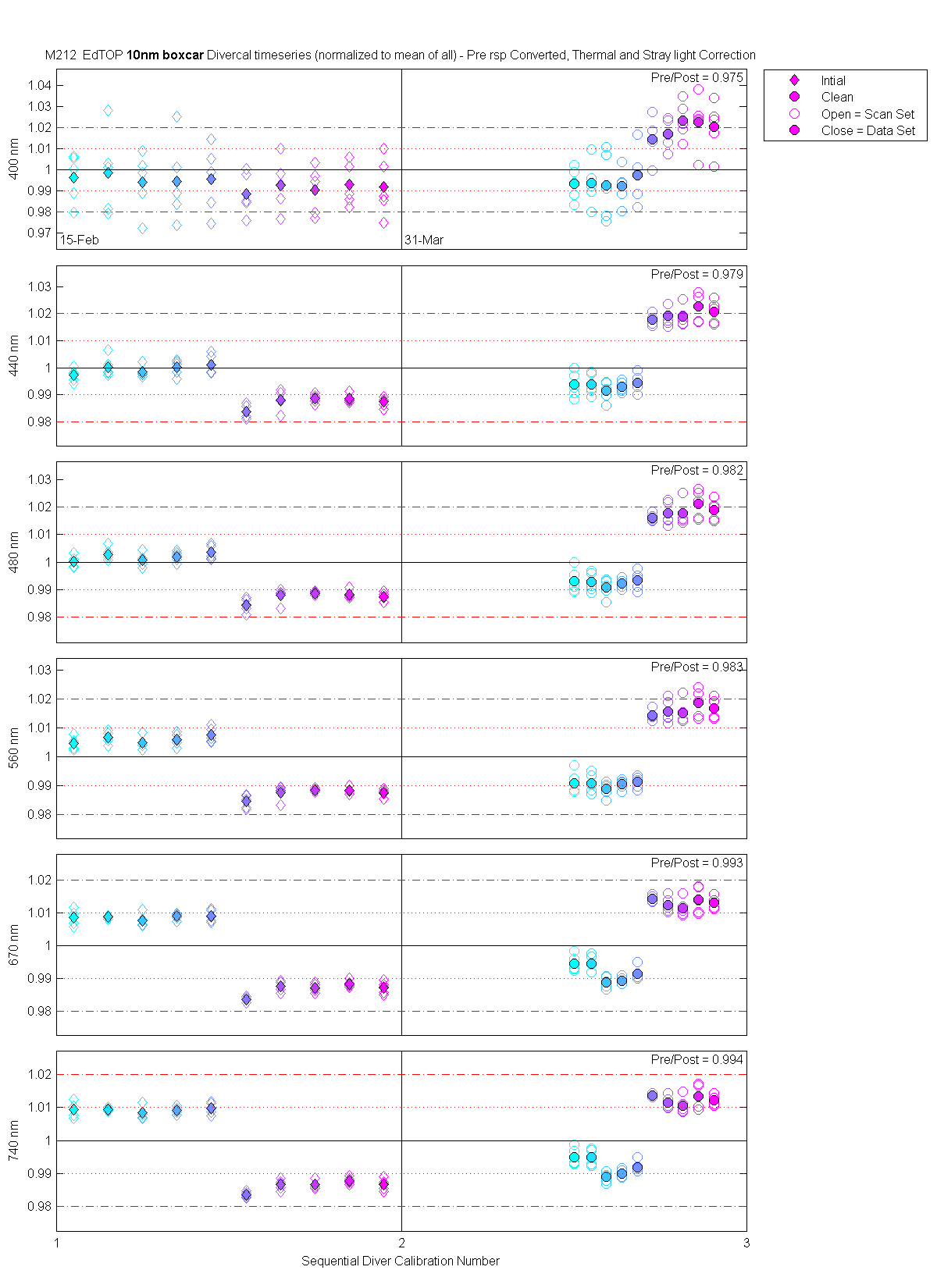
The scan sets usually consist of 5 light scans. A mean dark is subtracted off each light scan and they are divided by their intergration time and bin factor. The result in the graphs below are 5 light scans (the open symbols below). The solid symbol (same color) is the mean of all the good scans. Each scan sets were quality checked prior to processing. Each of the 5 lite scan were marked good, bad or wrong. Wrong means the data where collected when the diver cal lamps were not on the arm (so the data where in-water) or the lamp was not turned on (so only darks were collected). The bad data where any cal lamp light scans which stood out from the other scans as to variable. I was looking for scan sets with a percent standard deviation greater than 0.5%. If I had to remove more than 2 scan to get the %std below 0.5% then I removed no scans and left the data set intact. I then took all the good diver cal data for the entire deployment and calculated a mean. All the data plotted and in the tables are normalized to the mean of all the data (for the Sensor and 10 nm wide boxcar average).
The 10 nm boxcar bands were used to weight the data. In the plots the diamonds are the intial cal (beginning of the deploy, if they exist). The stars are the dirty cals and the circles are the clean cals (if they exist). The x symbols mark data sets where we dont know if it is intital, clean or dirty. Bad data are red symbols and wrong data are black. The graphs are scaled to the good data only so alot of the bad and wrong data sets do not show. The calibration number is the x-axis this number is also in the table under "Plot Number" so you can match up the table data with the plot data. The red horitontal lines show the 1, 2 and 5% lines The dates the divercal files were collected are listing at the bottom of the first axes in each figure.
Quick links to the graphs and table for each sensor.
edtop edmid edbot lutop lumid lubot
| MOBY | Cal | Cal | Data | Plot | Number | Mean of good divided by mean of all / %std | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensor | Date | file | Number | Type | Set# | Number | Good | 400 nm | 440 nm | 480 nm | 560 nm | 670 nm | 740 nm |
| EdTop | 15-Feb-2000 01:09:42 | 00021500 | 1 | Intial | 27 | 1.05 | 5 | 0.996 / 1.16 | 0.997 / 0.24 | 1.000 / 0.21 | 1.005 / 0.21 | 1.009 / 0.23 | 1.009 / 0.22 |
| EdTop | 15-Feb-2000 01:10:36 | 00021500 | 1 | Intial | 28 | 1.15 | 5 | 0.999 / 1.99 | 1.000 / 0.38 | 1.003 / 0.25 | 1.007 / 0.21 | 1.009 / 0.04 | 1.009 / 0.05 |
| EdTop | 15-Feb-2000 01:11:31 | 00021500 | 1 | Intial | 29 | 1.25 | 5 | 0.994 / 1.42 | 0.998 / 0.22 | 1.001 / 0.24 | 1.005 / 0.21 | 1.008 / 0.20 | 1.008 / 0.19 |
| EdTop | 15-Feb-2000 01:12:22 | 00021500 | 1 | Intial | 30 | 1.35 | 5 | 0.994 / 1.99 | 1.000 / 0.29 | 1.002 / 0.20 | 1.006 / 0.21 | 1.009 / 0.09 | 1.009 / 0.10 |
| EdTop | 15-Feb-2000 01:13:18 | 00021500 | 1 | Intial | 31 | 1.45 | 5 | 0.995 / 1.61 | 1.001 / 0.37 | 1.003 / 0.27 | 1.008 / 0.26 | 1.009 / 0.18 | 1.010 / 0.18 |
| EdTop | 15-Feb-2000 01:46:47 | 00021501 | 1 | Intial | 11 | 1.55 | 5 | 0.989 / 1.03 | 0.984 / 0.24 | 0.984 / 0.26 | 0.985 / 0.24 | 0.984 / 0.09 | 0.984 / 0.09 |
| EdTop | 15-Feb-2000 01:47:39 | 00021501 | 1 | Intial | 12 | 1.65 | 5 | 0.993 / 1.27 | 0.988 / 0.38 | 0.988 / 0.29 | 0.988 / 0.25 | 0.988 / 0.16 | 0.987 / 0.16 |
| EdTop | 15-Feb-2000 01:48:32 | 00021501 | 1 | Intial | 13 | 1.75 | 5 | 0.990 / 1.15 | 0.989 / 0.16 | 0.989 / 0.06 | 0.988 / 0.07 | 0.987 / 0.13 | 0.987 / 0.12 |
| EdTop | 15-Feb-2000 01:49:26 | 00021501 | 1 | Intial | 14 | 1.85 | 5 | 0.993 / 1.03 | 0.989 / 0.16 | 0.988 / 0.14 | 0.988 / 0.11 | 0.988 / 0.10 | 0.988 / 0.10 |
| EdTop | 15-Feb-2000 01:50:23 | 00021501 | 1 | Intial | 15 | 1.95 | 5 | 0.992 / 1.40 | 0.988 / 0.20 | 0.988 / 0.15 | 0.988 / 0.13 | 0.987 / 0.19 | 0.987 / 0.18 |
| EdTop | 31-Mar-2000 20:26:50 | 00033119 | 2 | Clean | 28 | 2.50 | 5 | 0.993 / 0.78 | 0.994 / 0.48 | 0.993 / 0.46 | 0.991 / 0.39 | 0.995 / 0.25 | 0.995 / 0.26 |
| EdTop | 31-Mar-2000 20:27:45 | 00033119 | 2 | Clean | 29 | 2.55 | 5 | 0.993 / 1.09 | 0.994 / 0.40 | 0.993 / 0.36 | 0.991 / 0.34 | 0.994 / 0.27 | 0.995 / 0.24 |
| EdTop | 31-Mar-2000 20:28:43 | 00033119 | 2 | Clean | 30 | 2.59 | 5 | 0.992 / 1.63 | 0.991 / 0.36 | 0.991 / 0.34 | 0.989 / 0.26 | 0.989 / 0.17 | 0.989 / 0.17 |
| EdTop | 31-Mar-2000 20:29:37 | 00033119 | 2 | Clean | 31 | 2.64 | 5 | 0.992 / 0.86 | 0.993 / 0.21 | 0.992 / 0.18 | 0.991 / 0.17 | 0.989 / 0.12 | 0.990 / 0.12 |
| EdTop | 31-Mar-2000 20:30:32 | 00033119 | 2 | Clean | 32 | 2.68 | 5 | 0.997 / 1.32 | 0.995 / 0.34 | 0.993 / 0.34 | 0.991 / 0.22 | 0.991 / 0.21 | 0.992 / 0.19 |
| EdTop | 31-Mar-2000 21:09:16 | 00033120 | 2 | Clean | 12 | 2.73 | 5 | 1.014 / 1.00 | 1.018 / 0.19 | 1.016 / 0.13 | 1.014 / 0.17 | 1.014 / 0.09 | 1.014 / 0.06 |
| EdTop | 31-Mar-2000 21:10:11 | 00033120 | 2 | Clean | 13 | 2.77 | 5 | 1.017 / 0.70 | 1.019 / 0.39 | 1.018 / 0.42 | 1.016 / 0.40 | 1.012 / 0.23 | 1.011 / 0.19 |
| EdTop | 31-Mar-2000 21:11:07 | 00033120 | 2 | Clean | 14 | 2.82 | 5 | 1.023 / 0.84 | 1.019 / 0.38 | 1.018 / 0.44 | 1.015 / 0.40 | 1.011 / 0.26 | 1.011 / 0.26 |
| EdTop | 31-Mar-2000 21:12:00 | 00033120 | 2 | Clean | 15 | 2.86 | 5 | 1.023 / 1.26 | 1.023 / 0.52 | 1.021 / 0.50 | 1.019 / 0.46 | 1.014 / 0.39 | 1.013 / 0.36 |
| EdTop | 31-Mar-2000 21:12:54 | 00033120 | 2 | Clean | 16 | 2.91 | 5 | 1.020 / 1.19 | 1.021 / 0.42 | 1.019 / 0.36 | 1.017 / 0.33 | 1.013 / 0.18 | 1.012 / 0.15 |
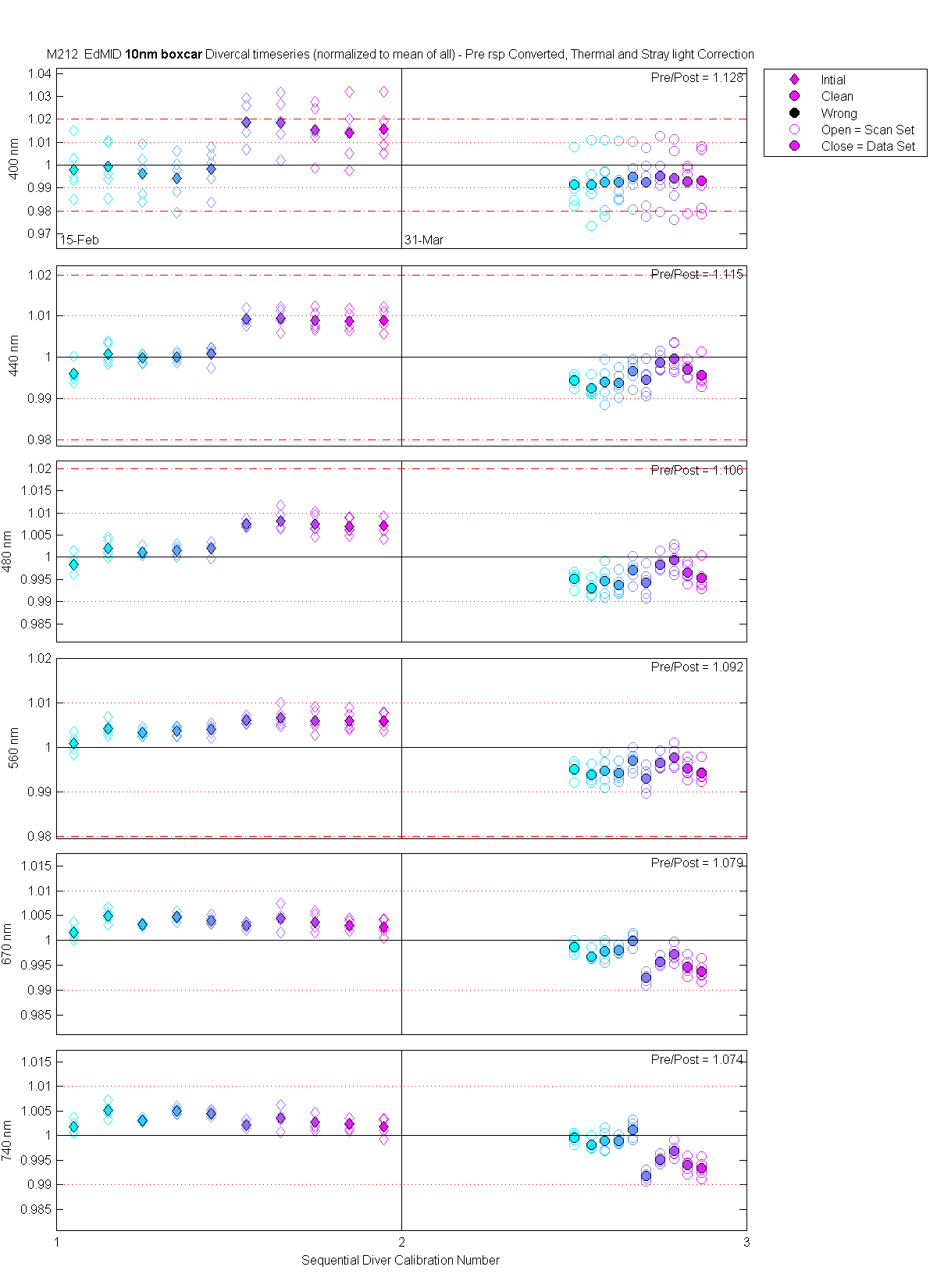
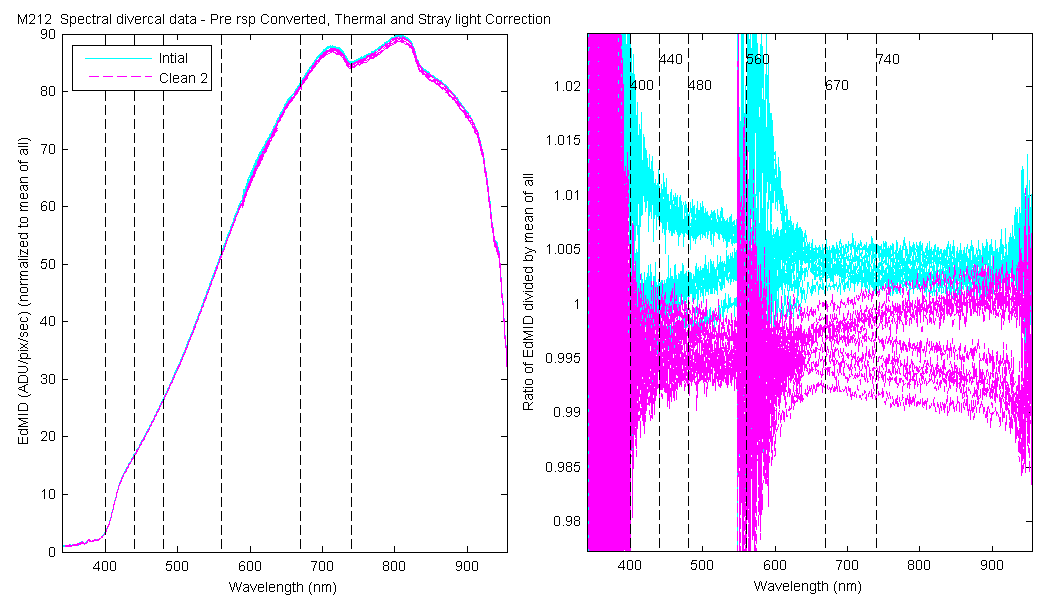
| MOBY | Cal | Cal | Data | Plot | Number | Mean of good divided by mean of all / %std | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensor | Date | file | Number | Type | Set# | Number | Good | 400 nm | 440 nm | 480 nm | 560 nm | 670 nm | 740 nm |
| EdMid | 15-Feb-2000 01:03:45 | 00021500 | 1 | Intial | 22 | 1.05 | 5 | 0.998 / 1.14 | 0.996 / 0.24 | 0.998 / 0.23 | 1.001 / 0.20 | 1.002 / 0.14 | 1.002 / 0.13 |
| EdMid | 15-Feb-2000 01:04:39 | 00021500 | 1 | Intial | 23 | 1.15 | 5 | 0.999 / 1.08 | 1.001 / 0.26 | 1.002 / 0.20 | 1.004 / 0.16 | 1.005 / 0.13 | 1.005 / 0.15 |
| EdMid | 15-Feb-2000 01:05:33 | 00021500 | 1 | Intial | 24 | 1.25 | 5 | 0.996 / 1.06 | 1.000 / 0.11 | 1.001 / 0.09 | 1.003 / 0.08 | 1.003 / 0.03 | 1.003 / 0.04 |
| EdMid | 15-Feb-2000 01:06:26 | 00021500 | 1 | Intial | 25 | 1.35 | 5 | 0.994 / 1.07 | 1.000 / 0.11 | 1.002 / 0.11 | 1.004 / 0.11 | 1.005 / 0.08 | 1.005 / 0.06 |
| EdMid | 15-Feb-2000 01:07:18 | 00021500 | 1 | Intial | 26 | 1.45 | 5 | 0.998 / 0.96 | 1.001 / 0.19 | 1.002 / 0.13 | 1.004 / 0.13 | 1.004 / 0.08 | 1.005 / 0.05 |
| EdMid | 15-Feb-2000 01:40:56 | 00021501 | 1 | Intial | 6 | 1.55 | 5 | 1.019 / 0.89 | 1.009 / 0.16 | 1.007 / 0.08 | 1.006 / 0.08 | 1.003 / 0.07 | 1.002 / 0.07 |
| EdMid | 15-Feb-2000 01:41:50 | 00021501 | 1 | Intial | 7 | 1.65 | 5 | 1.019 / 1.13 | 1.010 / 0.24 | 1.008 / 0.23 | 1.007 / 0.21 | 1.004 / 0.21 | 1.003 / 0.20 |
| EdMid | 15-Feb-2000 01:42:44 | 00021501 | 1 | Intial | 8 | 1.75 | 5 | 1.015 / 1.13 | 1.009 / 0.24 | 1.007 / 0.24 | 1.006 / 0.25 | 1.004 / 0.18 | 1.003 / 0.17 |
| EdMid | 15-Feb-2000 01:43:39 | 00021501 | 1 | Intial | 9 | 1.85 | 5 | 1.014 / 1.32 | 1.009 / 0.23 | 1.007 / 0.19 | 1.006 / 0.20 | 1.003 / 0.11 | 1.002 / 0.12 |
| EdMid | 15-Feb-2000 01:44:39 | 00021501 | 1 | Intial | 10 | 1.95 | 5 | 1.016 / 1.03 | 1.009 / 0.26 | 1.007 / 0.22 | 1.006 / 0.18 | 1.003 / 0.16 | 1.002 / 0.17 |
| EdMid | 31-Mar-2000 20:20:01 | 00033119 | 2 | Clean | 23 | 2.50 | 5 | 0.992 / 1.02 | 0.994 / 0.14 | 0.995 / 0.16 | 0.995 / 0.18 | 0.999 / 0.10 | 1.000 / 0.10 |
| EdMid | 31-Mar-2000 20:20:54 | 00033119 | 2 | Clean | 24 | 2.55 | 5 | 0.991 / 1.38 | 0.992 / 0.20 | 0.993 / 0.17 | 0.994 / 0.16 | 0.997 / 0.10 | 0.998 / 0.11 |
| EdMid | 31-Mar-2000 20:21:51 | 00033119 | 2 | Clean | 25 | 2.59 | 5 | 0.993 / 1.39 | 0.994 / 0.42 | 0.995 / 0.34 | 0.995 / 0.32 | 0.998 / 0.20 | 0.999 / 0.21 |
| EdMid | 31-Mar-2000 20:22:45 | 00033119 | 2 | Clean | 26 | 2.63 | 5 | 0.993 / 1.07 | 0.994 / 0.29 | 0.994 / 0.23 | 0.994 / 0.18 | 0.998 / 0.08 | 0.999 / 0.07 |
| EdMid | 31-Mar-2000 20:23:38 | 00033119 | 2 | Clean | 27 | 2.67 | 5 | 0.995 / 1.09 | 0.997 / 0.29 | 0.997 / 0.25 | 0.997 / 0.23 | 1.000 / 0.15 | 1.001 / 0.18 |
| EdMid | 31-Mar-2000 21:02:42 | 00033120 | 2 | Clean | 7 | 2.71 | 5 | 0.992 / 1.25 | 0.995 / 0.37 | 0.994 / 0.31 | 0.993 / 0.27 | 0.992 / 0.11 | 0.992 / 0.09 |
| EdMid | 31-Mar-2000 21:03:41 | 00033120 | 2 | Clean | 8 | 2.75 | 5 | 0.995 / 1.22 | 0.999 / 0.22 | 0.998 / 0.19 | 0.997 / 0.17 | 0.996 / 0.09 | 0.995 / 0.09 |
| EdMid | 31-Mar-2000 21:04:34 | 00033120 | 2 | Clean | 9 | 2.79 | 5 | 0.994 / 1.44 | 1.000 / 0.35 | 0.999 / 0.30 | 0.998 / 0.23 | 0.997 / 0.17 | 0.997 / 0.15 |
| EdMid | 31-Mar-2000 21:05:29 | 00033120 | 2 | Clean | 10 | 2.83 | 5 | 0.993 / 0.85 | 0.997 / 0.19 | 0.997 / 0.21 | 0.995 / 0.21 | 0.995 / 0.17 | 0.994 / 0.14 |
| EdMid | 31-Mar-2000 21:06:21 | 00033120 | 2 | Clean | 11 | 2.87 | 5 | 0.993 / 1.39 | 0.996 / 0.33 | 0.995 / 0.30 | 0.994 / 0.22 | 0.994 / 0.18 | 0.993 / 0.17 |
| EdMid | 31-Mar-2000 21:41:46 | 00033121 | 2 | Clean | 7 | 2.91 | 0 | NaN / NaN | NaN / NaN | NaN / NaN | NaN / NaN | NaN / NaN | NaN / NaN |
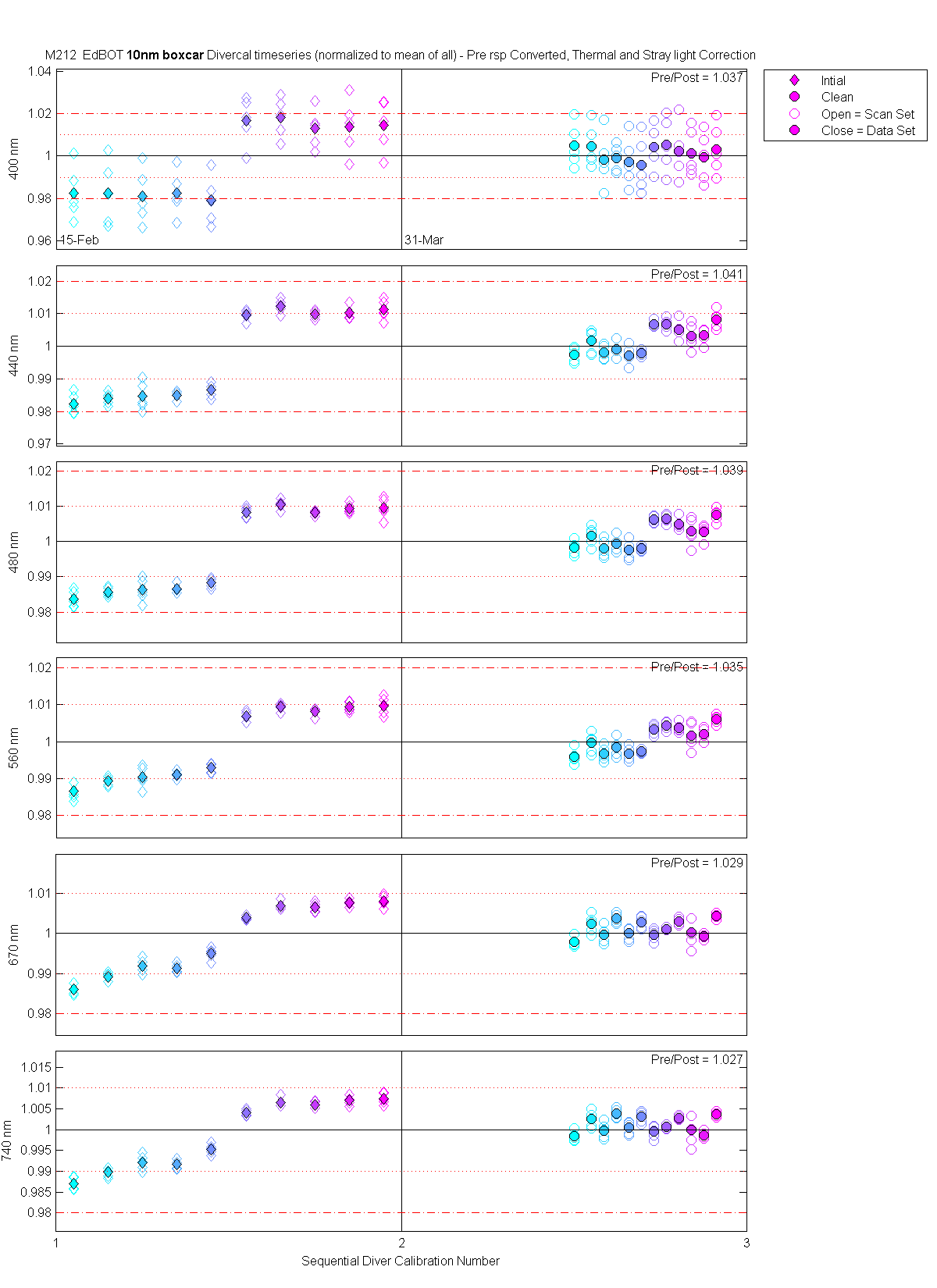
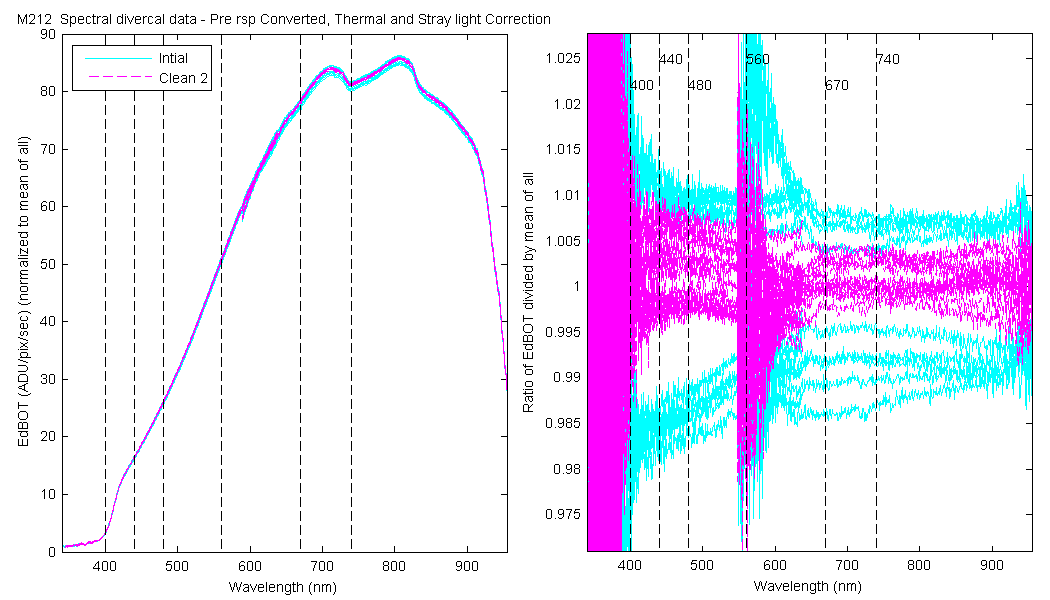
| MOBY | Cal | Cal | Data | Plot | Number | Mean of good divided by mean of all / %std | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensor | Date | file | Number | Type | Set# | Number | Good | 400 nm | 440 nm | 480 nm | 560 nm | 670 nm | 740 nm |
| EdBot | 15-Feb-2000 00:58:01 | 00021500 | 1 | Intial | 17 | 1.05 | 5 | 0.983 / 1.28 | 0.982 / 0.32 | 0.984 / 0.25 | 0.986 / 0.24 | 0.986 / 0.15 | 0.987 / 0.16 |
| EdBot | 15-Feb-2000 00:58:54 | 00021500 | 1 | Intial | 18 | 1.15 | 5 | 0.983 / 1.55 | 0.984 / 0.19 | 0.986 / 0.12 | 0.989 / 0.12 | 0.989 / 0.11 | 0.990 / 0.11 |
| EdBot | 15-Feb-2000 00:59:47 | 00021500 | 1 | Intial | 19 | 1.25 | 5 | 0.981 / 1.33 | 0.985 / 0.44 | 0.986 / 0.33 | 0.990 / 0.29 | 0.992 / 0.18 | 0.992 / 0.19 |
| EdBot | 15-Feb-2000 01:00:40 | 00021500 | 1 | Intial | 20 | 1.35 | 5 | 0.982 / 1.08 | 0.985 / 0.12 | 0.987 / 0.12 | 0.991 / 0.09 | 0.991 / 0.11 | 0.992 / 0.10 |
| EdBot | 15-Feb-2000 01:01:32 | 00021500 | 1 | Intial | 21 | 1.45 | 5 | 0.979 / 1.17 | 0.987 / 0.21 | 0.988 / 0.14 | 0.993 / 0.13 | 0.995 / 0.14 | 0.995 / 0.12 |
| EdBot | 15-Feb-2000 01:35:09 | 00021501 | 1 | Intial | 1 | 1.55 | 5 | 1.017 / 1.10 | 1.010 / 0.15 | 1.008 / 0.15 | 1.007 / 0.12 | 1.004 / 0.06 | 1.004 / 0.08 |
| EdBot | 15-Feb-2000 01:36:03 | 00021501 | 1 | Intial | 2 | 1.65 | 5 | 1.018 / 0.92 | 1.012 / 0.20 | 1.010 / 0.13 | 1.009 / 0.10 | 1.007 / 0.10 | 1.007 / 0.10 |
| EdBot | 15-Feb-2000 01:36:55 | 00021501 | 1 | Intial | 3 | 1.75 | 5 | 1.013 / 0.91 | 1.010 / 0.12 | 1.008 / 0.06 | 1.008 / 0.10 | 1.007 / 0.11 | 1.006 / 0.07 |
| EdBot | 15-Feb-2000 01:37:48 | 00021501 | 1 | Intial | 4 | 1.85 | 5 | 1.014 / 1.31 | 1.010 / 0.19 | 1.009 / 0.14 | 1.009 / 0.13 | 1.008 / 0.08 | 1.007 / 0.10 |
| EdBot | 15-Feb-2000 01:38:39 | 00021501 | 1 | Intial | 5 | 1.95 | 5 | 1.015 / 1.20 | 1.011 / 0.29 | 1.010 / 0.28 | 1.010 / 0.24 | 1.008 / 0.14 | 1.008 / 0.14 |
| EdBot | 31-Mar-2000 20:12:57 | 00033119 | 2 | Clean | 17 | 2.50 | 5 | 1.005 / 1.00 | 0.997 / 0.22 | 0.998 / 0.20 | 0.996 / 0.19 | 0.998 / 0.12 | 0.998 / 0.12 |
| EdBot | 31-Mar-2000 20:13:49 | 00033119 | 2 | Clean | 18 | 2.55 | 5 | 1.004 / 1.00 | 1.002 / 0.36 | 1.002 / 0.27 | 1.000 / 0.27 | 1.002 / 0.23 | 1.003 / 0.19 |
| EdBot | 31-Mar-2000 20:14:44 | 00033119 | 2 | Clean | 19 | 2.59 | 5 | 0.998 / 1.26 | 0.998 / 0.22 | 0.998 / 0.25 | 0.997 / 0.20 | 1.000 / 0.21 | 1.000 / 0.19 |
| EdBot | 31-Mar-2000 20:15:38 | 00033119 | 2 | Clean | 20 | 2.62 | 5 | 0.999 / 0.62 | 0.999 / 0.22 | 0.999 / 0.22 | 0.998 / 0.24 | 1.004 / 0.12 | 1.004 / 0.11 |
| EdBot | 31-Mar-2000 20:16:35 | 00033119 | 2 | Clean | 21 | 2.66 | 5 | 0.997 / 1.16 | 0.997 / 0.28 | 0.998 / 0.26 | 0.997 / 0.18 | 1.000 / 0.16 | 1.000 / 0.15 |
| EdBot | 31-Mar-2000 20:17:28 | 00033119 | 2 | Clean | 22 | 2.70 | 5 | 0.996 / 1.32 | 0.998 / 0.09 | 0.998 / 0.07 | 0.997 / 0.07 | 1.003 / 0.15 | 1.003 / 0.13 |
| EdBot | 31-Mar-2000 20:55:46 | 00033120 | 2 | Clean | 1 | 2.73 | 5 | 1.004 / 1.01 | 1.007 / 0.09 | 1.006 / 0.10 | 1.003 / 0.15 | 1.000 / 0.15 | 0.999 / 0.15 |
| EdBot | 31-Mar-2000 20:56:40 | 00033120 | 2 | Clean | 2 | 2.77 | 5 | 1.005 / 1.28 | 1.007 / 0.17 | 1.006 / 0.12 | 1.004 / 0.12 | 1.001 / 0.04 | 1.001 / 0.04 |
| EdBot | 31-Mar-2000 20:57:34 | 00033120 | 2 | Clean | 3 | 2.80 | 5 | 1.002 / 1.29 | 1.005 / 0.29 | 1.005 / 0.17 | 1.004 / 0.13 | 1.003 / 0.09 | 1.003 / 0.05 |
| EdBot | 31-Mar-2000 20:58:28 | 00033120 | 2 | Clean | 4 | 2.84 | 5 | 1.001 / 1.12 | 1.003 / 0.38 | 1.003 / 0.38 | 1.002 / 0.36 | 1.000 / 0.36 | 1.000 / 0.36 |
| EdBot | 31-Mar-2000 20:59:30 | 00033120 | 2 | Clean | 5 | 2.88 | 5 | 0.999 / 1.16 | 1.003 / 0.24 | 1.003 / 0.20 | 1.002 / 0.17 | 0.999 / 0.06 | 0.999 / 0.08 |
| EdBot | 31-Mar-2000 21:00:24 | 00033120 | 2 | Clean | 6 | 2.91 | 5 | 1.003 / 1.19 | 1.008 / 0.27 | 1.007 / 0.18 | 1.006 / 0.13 | 1.004 / 0.06 | 1.004 / 0.06 |
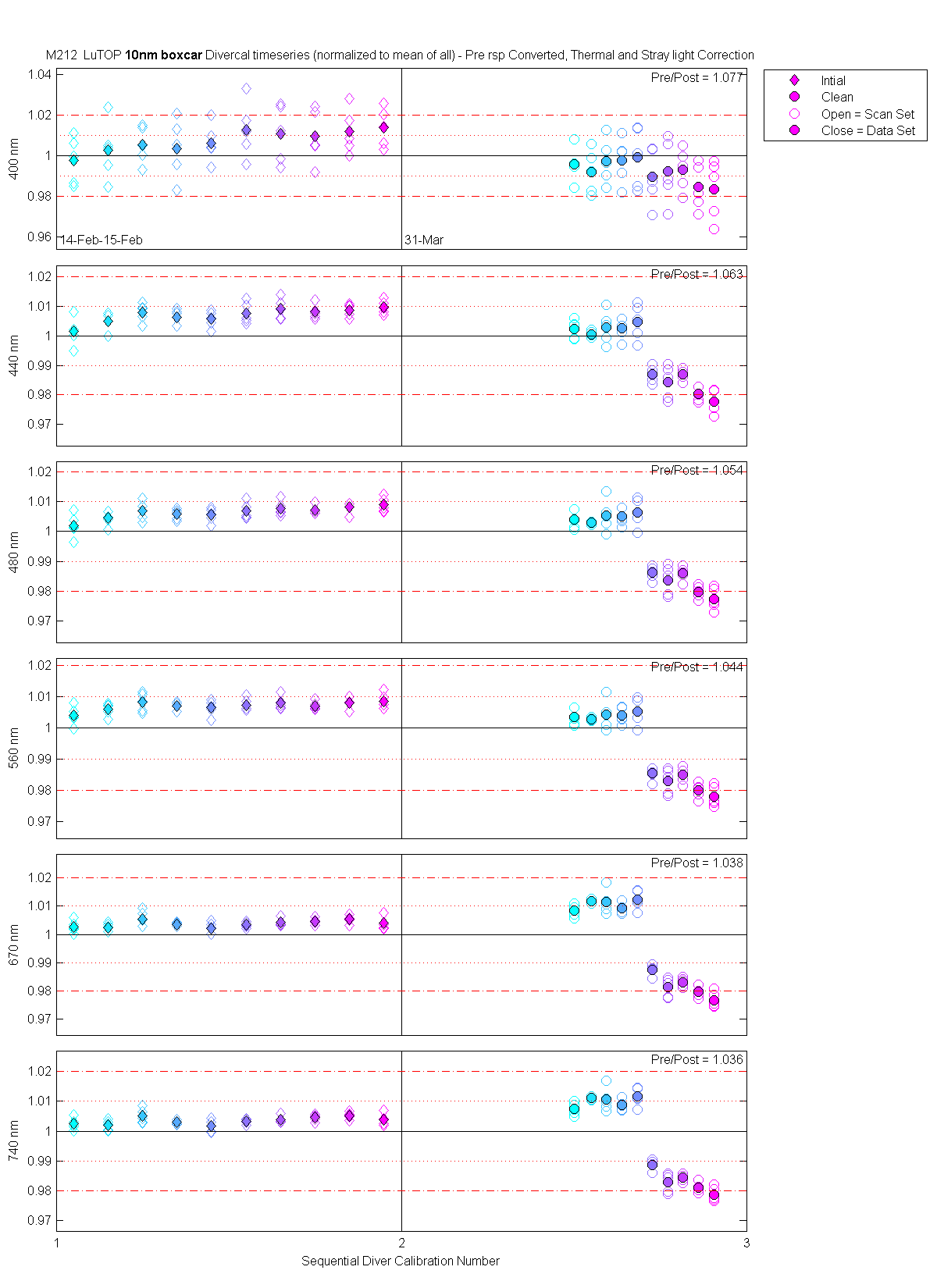
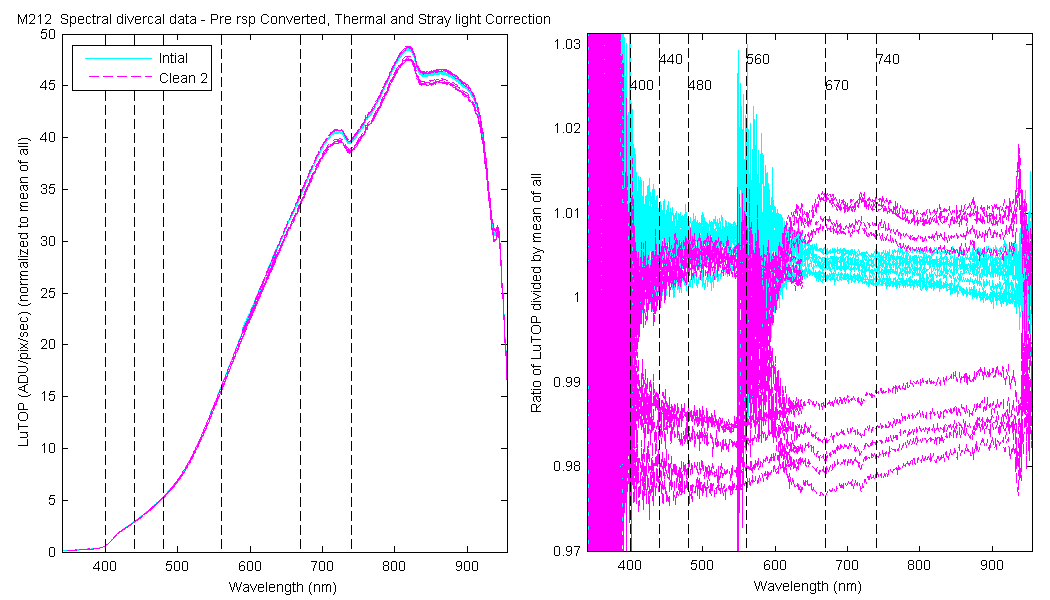
| MOBY | Cal | Cal | Data | Plot | Number | Mean of good divided by mean of all / %std | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensor | Date | file | Number | Type | Set# | Number | Good | 400 nm | 440 nm | 480 nm | 560 nm | 670 nm | 740 nm |
| LuTop | 14-Feb-2000 23:50:15 | 00021423 | 1 | Intial | 12 | 1.05 | 5 | 0.998 / 1.17 | 1.001 / 0.47 | 1.002 / 0.39 | 1.004 / 0.30 | 1.003 / 0.22 | 1.002 / 0.19 |
| LuTop | 14-Feb-2000 23:51:05 | 00021423 | 1 | Intial | 13 | 1.15 | 5 | 1.003 / 1.45 | 1.005 / 0.30 | 1.005 / 0.25 | 1.006 / 0.21 | 1.002 / 0.16 | 1.002 / 0.16 |
| LuTop | 14-Feb-2000 23:51:57 | 00021423 | 1 | Intial | 14 | 1.25 | 5 | 1.005 / 0.92 | 1.008 / 0.30 | 1.007 / 0.31 | 1.008 / 0.30 | 1.006 / 0.27 | 1.005 / 0.24 |
| LuTop | 14-Feb-2000 23:52:47 | 00021423 | 1 | Intial | 15 | 1.35 | 5 | 1.004 / 1.48 | 1.006 / 0.27 | 1.006 / 0.18 | 1.007 / 0.16 | 1.004 / 0.06 | 1.003 / 0.08 |
| LuTop | 14-Feb-2000 23:53:36 | 00021423 | 1 | Intial | 16 | 1.45 | 5 | 1.006 / 0.94 | 1.006 / 0.28 | 1.006 / 0.23 | 1.006 / 0.25 | 1.002 / 0.20 | 1.002 / 0.20 |
| LuTop | 15-Feb-2000 00:29:43 | 00021500 | 1 | Intial | 12 | 1.55 | 5 | 1.013 / 1.36 | 1.008 / 0.36 | 1.007 / 0.28 | 1.007 / 0.21 | 1.003 / 0.10 | 1.003 / 0.08 |
| LuTop | 15-Feb-2000 00:30:34 | 00021500 | 1 | Intial | 13 | 1.65 | 5 | 1.011 / 1.42 | 1.009 / 0.34 | 1.008 / 0.24 | 1.008 / 0.22 | 1.004 / 0.13 | 1.004 / 0.12 |
| LuTop | 15-Feb-2000 00:31:22 | 00021500 | 1 | Intial | 14 | 1.75 | 5 | 1.010 / 1.32 | 1.008 / 0.24 | 1.007 / 0.15 | 1.007 / 0.14 | 1.005 / 0.11 | 1.004 / 0.11 |
| LuTop | 15-Feb-2000 00:32:26 | 00021500 | 1 | Intial | 15 | 1.85 | 5 | 1.012 / 1.09 | 1.009 / 0.21 | 1.008 / 0.19 | 1.008 / 0.17 | 1.005 / 0.14 | 1.005 / 0.11 |
| LuTop | 15-Feb-2000 00:33:15 | 00021500 | 1 | Intial | 16 | 1.95 | 5 | 1.014 / 0.94 | 1.010 / 0.23 | 1.009 / 0.25 | 1.008 / 0.26 | 1.004 / 0.22 | 1.004 / 0.20 |
| LuTop | 31-Mar-2000 19:44:49 | 00033119 | 2 | Clean | 12 | 2.50 | 5 | 0.996 / 0.85 | 1.002 / 0.32 | 1.004 / 0.33 | 1.004 / 0.28 | 1.008 / 0.21 | 1.007 / 0.20 |
| LuTop | 31-Mar-2000 19:45:39 | 00033119 | 2 | Clean | 13 | 2.55 | 5 | 0.992 / 1.08 | 1.001 / 0.12 | 1.003 / 0.04 | 1.003 / 0.04 | 1.012 / 0.08 | 1.011 / 0.06 |
| LuTop | 31-Mar-2000 19:46:30 | 00033119 | 2 | Clean | 14 | 2.59 | 5 | 0.997 / 1.11 | 1.003 / 0.55 | 1.005 / 0.54 | 1.004 / 0.47 | 1.012 / 0.42 | 1.010 / 0.38 |
| LuTop | 31-Mar-2000 19:47:18 | 00033119 | 2 | Clean | 15 | 2.64 | 5 | 0.998 / 1.13 | 1.003 / 0.36 | 1.005 / 0.29 | 1.004 / 0.25 | 1.009 / 0.18 | 1.009 / 0.18 |
| LuTop | 31-Mar-2000 19:48:07 | 00033119 | 2 | Clean | 16 | 2.68 | 5 | 0.999 / 1.51 | 1.005 / 0.60 | 1.006 / 0.48 | 1.005 / 0.42 | 1.012 / 0.34 | 1.012 / 0.30 |
| LuTop | 31-Mar-2000 21:47:40 | 00033121 | 2 | Clean | 11 | 2.73 | 5 | 0.990 / 1.41 | 0.987 / 0.27 | 0.986 / 0.23 | 0.985 / 0.21 | 0.987 / 0.19 | 0.989 / 0.17 |
| LuTop | 31-Mar-2000 21:48:30 | 00033121 | 2 | Clean | 12 | 2.77 | 5 | 0.992 / 1.58 | 0.984 / 0.58 | 0.984 / 0.51 | 0.983 / 0.42 | 0.981 / 0.35 | 0.983 / 0.33 |
| LuTop | 31-Mar-2000 21:49:18 | 00033121 | 2 | Clean | 13 | 2.82 | 5 | 0.993 / 1.04 | 0.987 / 0.20 | 0.986 / 0.24 | 0.985 / 0.25 | 0.983 / 0.16 | 0.985 / 0.14 |
| LuTop | 31-Mar-2000 21:50:08 | 00033121 | 2 | Clean | 14 | 2.86 | 5 | 0.984 / 1.15 | 0.980 / 0.25 | 0.980 / 0.22 | 0.980 / 0.24 | 0.980 / 0.19 | 0.981 / 0.17 |
| LuTop | 31-Mar-2000 21:51:01 | 00033121 | 2 | Clean | 15 | 2.91 | 5 | 0.984 / 1.48 | 0.978 / 0.39 | 0.977 / 0.38 | 0.978 / 0.34 | 0.977 / 0.28 | 0.979 / 0.25 |
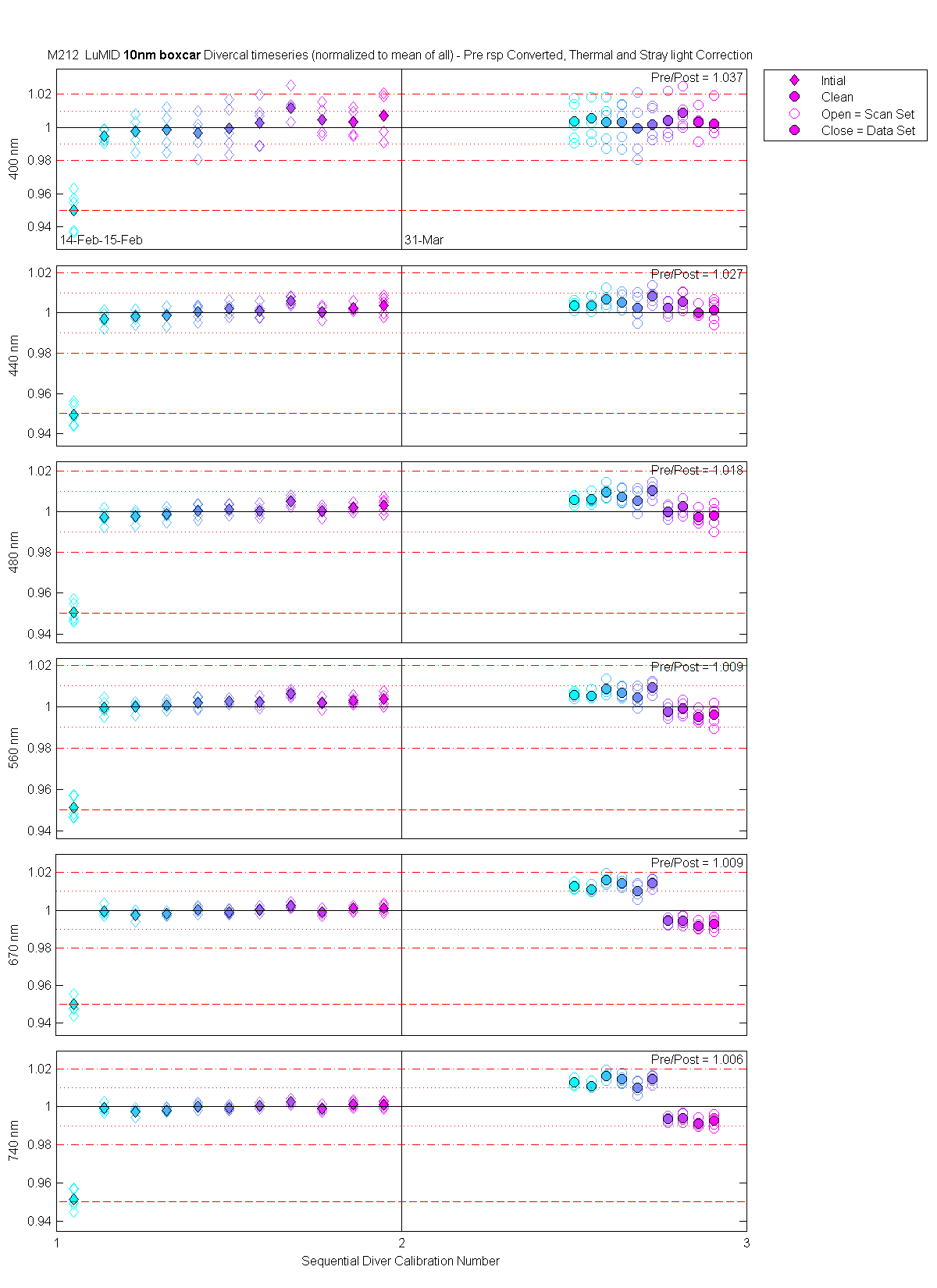
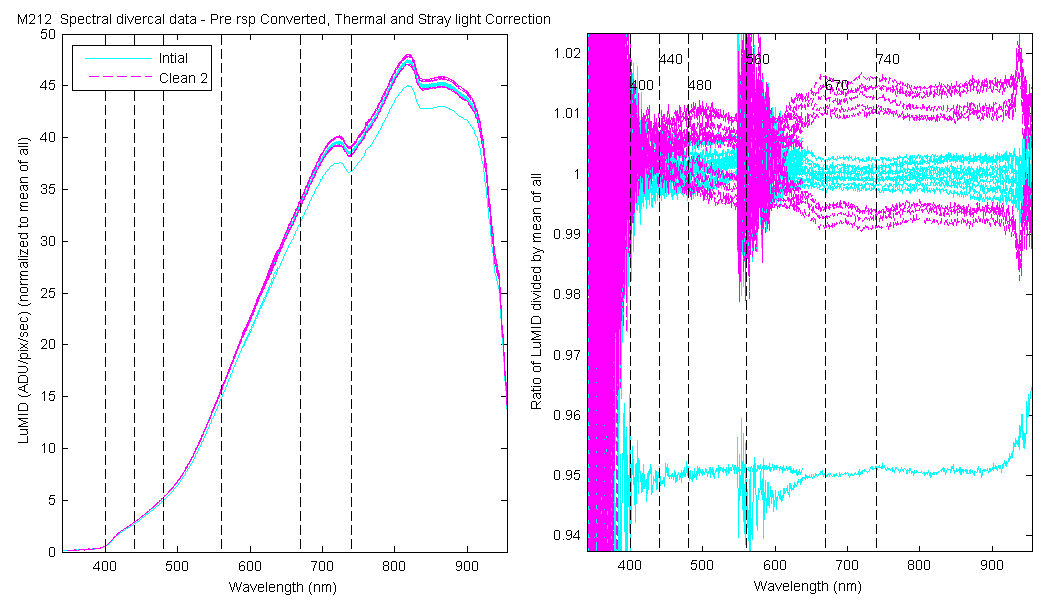
| MOBY | Cal | Cal | Data | Plot | Number | Mean of good divided by mean of all / %std | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensor | Date | file | Number | Type | Set# | Number | Good | 400 nm | 440 nm | 480 nm | 560 nm | 670 nm | 740 nm |
| LuMid | 14-Feb-2000 23:43:18 | 00021423 | 1 | Intial | 6 | 1.05 | 5 | 0.950 / 1.27 | 0.949 / 0.59 | 0.950 / 0.54 | 0.951 / 0.57 | 0.950 / 0.55 | 0.951 / 0.56 |
| LuMid | 14-Feb-2000 23:44:17 | 00021423 | 1 | Intial | 7 | 1.14 | 5 | 0.995 / 0.40 | 0.997 / 0.36 | 0.997 / 0.36 | 1.000 / 0.36 | 0.999 / 0.25 | 0.999 / 0.22 |
| LuMid | 14-Feb-2000 23:45:14 | 00021423 | 1 | Intial | 8 | 1.23 | 5 | 0.997 / 0.91 | 0.998 / 0.28 | 0.998 / 0.28 | 1.000 / 0.24 | 0.998 / 0.21 | 0.997 / 0.19 |
| LuMid | 14-Feb-2000 23:46:10 | 00021423 | 1 | Intial | 9 | 1.32 | 5 | 0.998 / 1.11 | 0.999 / 0.36 | 0.999 / 0.28 | 1.001 / 0.20 | 0.998 / 0.13 | 0.998 / 0.12 |
| LuMid | 14-Feb-2000 23:47:10 | 00021423 | 1 | Intial | 10 | 1.41 | 5 | 0.997 / 1.11 | 1.001 / 0.38 | 1.000 / 0.34 | 1.002 / 0.29 | 1.000 / 0.18 | 1.000 / 0.17 |
| LuMid | 14-Feb-2000 23:48:00 | 00021423 | 1 | Intial | 11 | 1.50 | 5 | 1.000 / 1.39 | 1.002 / 0.33 | 1.001 / 0.23 | 1.003 / 0.15 | 0.999 / 0.13 | 0.999 / 0.14 |
| LuMid | 15-Feb-2000 00:23:39 | 00021500 | 1 | Intial | 7 | 1.59 | 5 | 1.003 / 1.34 | 1.001 / 0.35 | 1.000 / 0.28 | 1.002 / 0.23 | 1.000 / 0.15 | 1.000 / 0.12 |
| LuMid | 15-Feb-2000 00:24:31 | 00021500 | 1 | Intial | 8 | 1.68 | 5 | 1.012 / 0.92 | 1.006 / 0.22 | 1.005 / 0.23 | 1.006 / 0.16 | 1.002 / 0.13 | 1.002 / 0.13 |
| LuMid | 15-Feb-2000 00:25:21 | 00021500 | 1 | Intial | 9 | 1.77 | 5 | 1.005 / 0.86 | 1.001 / 0.29 | 1.000 / 0.24 | 1.002 / 0.23 | 0.999 / 0.16 | 0.999 / 0.15 |
| LuMid | 15-Feb-2000 00:26:08 | 00021500 | 1 | Intial | 10 | 1.86 | 5 | 1.003 / 0.79 | 1.003 / 0.20 | 1.002 / 0.18 | 1.003 / 0.18 | 1.001 / 0.16 | 1.001 / 0.16 |
| LuMid | 15-Feb-2000 00:27:02 | 00021500 | 1 | Intial | 11 | 1.95 | 5 | 1.007 / 1.28 | 1.004 / 0.46 | 1.003 / 0.34 | 1.004 / 0.30 | 1.001 / 0.20 | 1.001 / 0.17 |
| LuMid | 31-Mar-2000 19:38:06 | 00033119 | 2 | Clean | 6 | 2.50 | 5 | 1.004 / 1.20 | 1.004 / 0.20 | 1.006 / 0.20 | 1.006 / 0.14 | 1.013 / 0.17 | 1.013 / 0.17 |
| LuMid | 31-Mar-2000 19:38:58 | 00033119 | 2 | Clean | 7 | 2.55 | 5 | 1.005 / 1.23 | 1.004 / 0.29 | 1.006 / 0.26 | 1.005 / 0.18 | 1.011 / 0.16 | 1.011 / 0.15 |
| LuMid | 31-Mar-2000 19:39:48 | 00033119 | 2 | Clean | 8 | 2.59 | 5 | 1.003 / 1.26 | 1.007 / 0.38 | 1.010 / 0.32 | 1.009 / 0.30 | 1.016 / 0.24 | 1.016 / 0.23 |
| LuMid | 31-Mar-2000 19:40:39 | 00033119 | 2 | Clean | 9 | 2.64 | 5 | 1.003 / 1.21 | 1.005 / 0.43 | 1.008 / 0.38 | 1.007 / 0.29 | 1.014 / 0.25 | 1.014 / 0.23 |
| LuMid | 31-Mar-2000 19:41:30 | 00033119 | 2 | Clean | 10 | 2.68 | 5 | 0.999 / 1.63 | 1.002 / 0.64 | 1.005 / 0.52 | 1.004 / 0.46 | 1.010 / 0.36 | 1.010 / 0.33 |
| LuMid | 31-Mar-2000 19:42:24 | 00033119 | 2 | Clean | 11 | 2.73 | 5 | 1.002 / 0.98 | 1.008 / 0.38 | 1.010 / 0.34 | 1.009 / 0.29 | 1.015 / 0.24 | 1.015 / 0.22 |
| LuMid | 31-Mar-2000 21:40:39 | 00033121 | 2 | Clean | 6 | 2.77 | 5 | 1.004 / 1.09 | 1.002 / 0.34 | 1.000 / 0.31 | 0.998 / 0.29 | 0.994 / 0.20 | 0.994 / 0.18 |
| LuMid | 31-Mar-2000 21:42:59 | 00033121 | 2 | Clean | 8 | 2.82 | 5 | 1.009 / 0.98 | 1.006 / 0.46 | 1.003 / 0.39 | 0.999 / 0.33 | 0.995 / 0.25 | 0.994 / 0.23 |
| LuMid | 31-Mar-2000 21:43:51 | 00033121 | 2 | Clean | 9 | 2.86 | 5 | 1.003 / 0.77 | 1.000 / 0.26 | 0.997 / 0.31 | 0.995 / 0.29 | 0.991 / 0.21 | 0.991 / 0.21 |
| LuMid | 31-Mar-2000 21:45:03 | 00033121 | 2 | Clean | 10 | 2.91 | 5 | 1.002 / 0.96 | 1.001 / 0.57 | 0.998 / 0.57 | 0.996 / 0.49 | 0.993 / 0.33 | 0.993 / 0.29 |
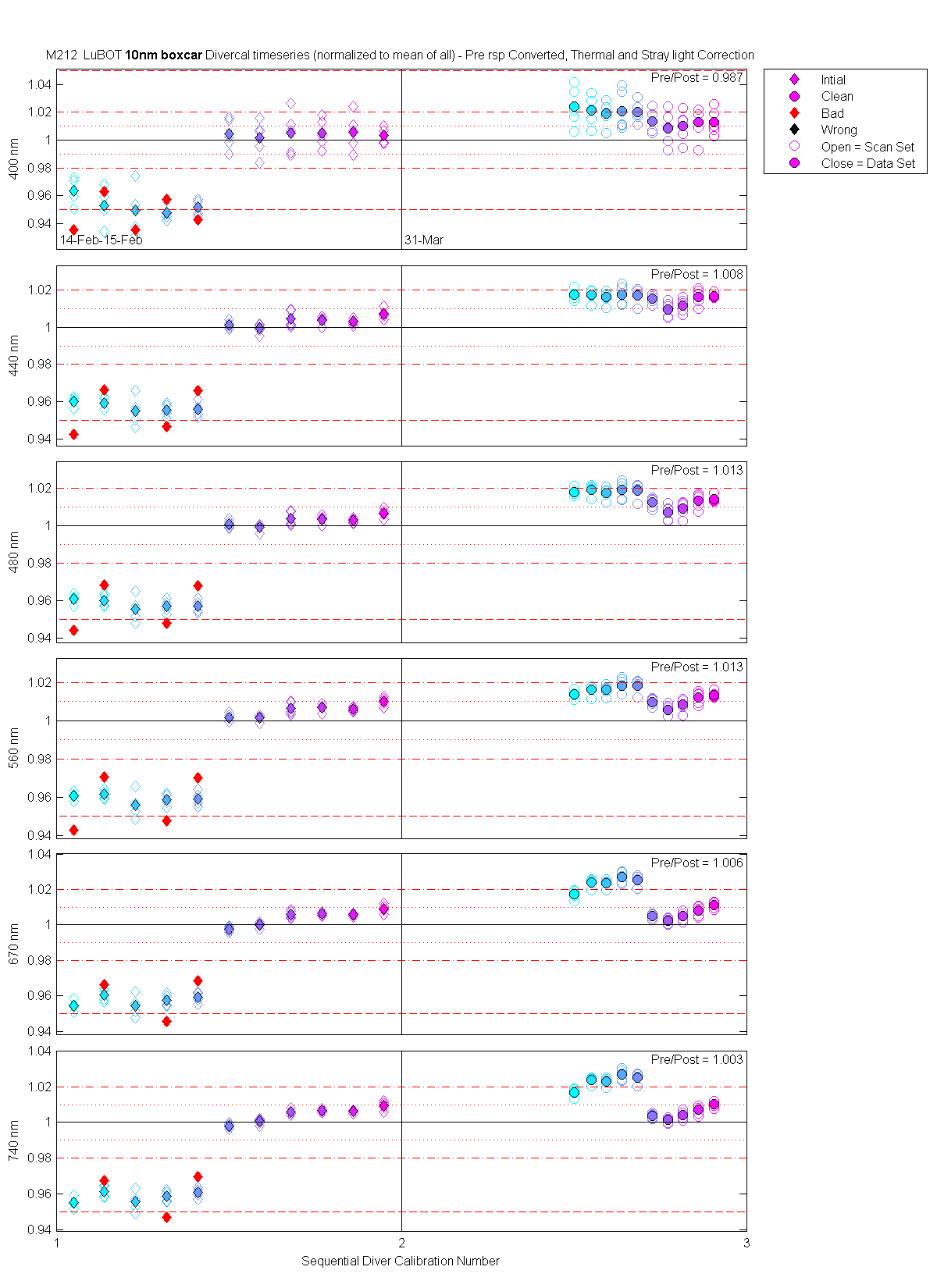
| MOBY | Cal | Cal | Data | Plot | Number | Mean of good divided by mean of all / %std | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensor | Date | file | Number | Type | Set# | Number | Good | 400 nm | 440 nm | 480 nm | 560 nm | 670 nm | 740 nm |
| LuBot | 14-Feb-2000 23:36:41 | 00021423 | 1 | Intial | 1 | 1.05 | 4 | 0.964 / 1.07 | 0.960 / 0.30 | 0.961 / 0.30 | 0.961 / 0.24 | 0.954 / 0.34 | 0.955 / 0.32 |
| LuBot | 14-Feb-2000 23:37:30 | 00021423 | 1 | Intial | 2 | 1.14 | 4 | 0.953 / 1.51 | 0.959 / 0.39 | 0.960 / 0.34 | 0.961 / 0.25 | 0.960 / 0.39 | 0.961 / 0.37 |
| LuBot | 14-Feb-2000 23:38:18 | 00021423 | 1 | Intial | 3 | 1.23 | 4 | 0.949 / 2.02 | 0.955 / 0.89 | 0.956 / 0.79 | 0.956 / 0.78 | 0.954 / 0.65 | 0.956 / 0.63 |
| LuBot | 14-Feb-2000 23:39:06 | 00021423 | 1 | Intial | 4 | 1.32 | 4 | 0.948 / 0.65 | 0.956 / 0.39 | 0.957 / 0.41 | 0.959 / 0.36 | 0.958 / 0.36 | 0.959 / 0.36 |
| LuBot | 14-Feb-2000 23:39:55 | 00021423 | 1 | Intial | 5 | 1.41 | 4 | 0.951 / 0.61 | 0.956 / 0.43 | 0.957 / 0.38 | 0.959 / 0.41 | 0.959 / 0.33 | 0.961 / 0.31 |
| LuBot | 15-Feb-2000 00:17:34 | 00021500 | 1 | Intial | 1 | 1.50 | 5 | 1.004 / 1.12 | 1.001 / 0.20 | 1.001 / 0.21 | 1.002 / 0.21 | 0.998 / 0.15 | 0.998 / 0.13 |
| LuBot | 15-Feb-2000 00:18:22 | 00021500 | 1 | Intial | 2 | 1.59 | 5 | 1.002 / 1.23 | 0.999 / 0.25 | 0.999 / 0.18 | 1.001 / 0.15 | 1.000 / 0.17 | 1.000 / 0.16 |
| LuBot | 15-Feb-2000 00:19:18 | 00021500 | 1 | Intial | 3 | 1.68 | 5 | 1.005 / 1.53 | 1.004 / 0.45 | 1.004 / 0.37 | 1.006 / 0.33 | 1.006 / 0.22 | 1.006 / 0.22 |
| LuBot | 15-Feb-2000 00:20:07 | 00021500 | 1 | Intial | 4 | 1.77 | 5 | 1.005 / 1.05 | 1.004 / 0.23 | 1.004 / 0.21 | 1.007 / 0.19 | 1.006 / 0.10 | 1.007 / 0.09 |
| LuBot | 15-Feb-2000 00:21:11 | 00021500 | 1 | Intial | 5 | 1.86 | 5 | 1.006 / 1.32 | 1.003 / 0.19 | 1.003 / 0.13 | 1.006 / 0.11 | 1.006 / 0.07 | 1.006 / 0.07 |
| LuBot | 15-Feb-2000 00:21:59 | 00021500 | 1 | Intial | 6 | 1.95 | 4 | 1.003 / 0.62 | 1.007 / 0.29 | 1.007 / 0.27 | 1.010 / 0.24 | 1.009 / 0.26 | 1.009 / 0.26 |
| LuBot | 31-Mar-2000 19:32:33 | 00033119 | 2 | Clean | 1 | 2.50 | 5 | 1.024 / 1.37 | 1.017 / 0.27 | 1.018 / 0.20 | 1.014 / 0.20 | 1.017 / 0.24 | 1.017 / 0.22 |
| LuBot | 31-Mar-2000 19:33:24 | 00033119 | 2 | Clean | 2 | 2.55 | 5 | 1.021 / 1.04 | 1.017 / 0.33 | 1.019 / 0.29 | 1.016 / 0.27 | 1.024 / 0.23 | 1.024 / 0.20 |
| LuBot | 31-Mar-2000 19:34:14 | 00033119 | 2 | Clean | 3 | 2.59 | 5 | 1.019 / 0.87 | 1.016 / 0.35 | 1.018 / 0.31 | 1.016 / 0.28 | 1.024 / 0.23 | 1.023 / 0.20 |
| LuBot | 31-Mar-2000 19:35:04 | 00033119 | 2 | Clean | 4 | 2.64 | 5 | 1.021 / 1.44 | 1.017 / 0.49 | 1.019 / 0.47 | 1.018 / 0.42 | 1.027 / 0.31 | 1.027 / 0.30 |
| LuBot | 31-Mar-2000 19:35:53 | 00033119 | 2 | Clean | 5 | 2.68 | 5 | 1.020 / 0.73 | 1.017 / 0.42 | 1.019 / 0.40 | 1.018 / 0.34 | 1.025 / 0.29 | 1.025 / 0.27 |
| LuBot | 31-Mar-2000 21:34:57 | 00033121 | 2 | Clean | 1 | 2.73 | 5 | 1.014 / 0.80 | 1.015 / 0.26 | 1.012 / 0.29 | 1.010 / 0.21 | 1.005 / 0.15 | 1.004 / 0.15 |
| LuBot | 31-Mar-2000 21:35:47 | 00033121 | 2 | Clean | 2 | 2.77 | 5 | 1.008 / 1.25 | 1.010 / 0.42 | 1.007 / 0.40 | 1.006 / 0.31 | 1.002 / 0.18 | 1.001 / 0.17 |
| LuBot | 31-Mar-2000 21:36:36 | 00033121 | 2 | Clean | 3 | 2.82 | 5 | 1.010 / 1.08 | 1.012 / 0.38 | 1.009 / 0.41 | 1.008 / 0.35 | 1.005 / 0.26 | 1.004 / 0.23 |
| LuBot | 31-Mar-2000 21:37:26 | 00033121 | 2 | Clean | 4 | 2.86 | 5 | 1.013 / 1.23 | 1.016 / 0.43 | 1.014 / 0.41 | 1.012 / 0.32 | 1.008 / 0.30 | 1.007 / 0.27 |
| LuBot | 31-Mar-2000 21:38:15 | 00033121 | 2 | Clean | 5 | 2.91 | 5 | 1.013 / 0.91 | 1.017 / 0.14 | 1.014 / 0.18 | 1.014 / 0.16 | 1.011 / 0.19 | 1.010 / 0.18 |
Plots of the auxilliary data for each sensor.